Welcome back to our “Decoding ‘Good’ AI” blog series, where we’re using real-world examples from the development of our own AI-powered oocyte assessment model to illustrate the key steps required to develop a high-quality machine learning model for medical image analysis.
In our previous blog post, we introduced you to the exciting world of machine learning and its practical application in reproductive medicine. We discussed the importance of clearly defining the specific task that the model completes and the essential considerations for building a robust dataset that enables the model to perform well in real-world clinical scenarios.
As we continue our series, we dive deeper into the development of a deep learning model for oocyte analysis. In this article, we’ll explore the next critical step: Choosing a deep learning model architecture.
Many people are interacting with AI on a daily basis now, but different tasks require AI models that are developed using different types of architectures and design. Here, we’ll explore best-in-class and emerging architectures for medical image analysis.
Stay tuned for our next blog post where we’ll dig into the process of training a deep learning model and considerations for how it’s trained. Later, we’ll close out the series by uncovering the final steps of our AI journey: evaluating the model’s performance and the continuous process of performance monitoring.
By the end of the series, clinic stakeholders will have a solid understanding of the fundamentals of ‘Good’ AI and should be able to assess the quality of AI models and their potential impact on clinical practice.
CHOOSING SUITABLE DEEP LEARNING MODEL ARCHITECTURES TO BUILD AN ENSEMBLE MODEL
Our last post discussed the importance of first defining a specific task for the model to complete to ensure that its architecture and design are selected to best address that challenge. As we approach this next section, recall that our model’s task is to classify oocyte images to predict whether they will form a blastocyst or not.
There are several types of model architectures that help to solve this, so part of our model development process involves experimenting with different architecture types to see which performs the strongest in making blastocyst predictions from oocyte images. We also experiment to see if multiple architecture types can be combined to work together to produce stronger predictive power as an ensemble model.
To help you understand the various architecture options available for our model development, let’s go on a quick history lesson.
Early image classification models often relied on handcrafted features extracted from images, such as texture, colour, or shape descriptors. These features were then fed into traditional machine learning algorithms such as Random Forest or XGBoost (both built using ensembles of decision trees) to classify the images. While these shallow architectures had some success with more basic classification tasks, they required manual feature identification and struggled to capture complex image patterns. Most importantly, the user needed to be able to identify these features themselves, which in many cases are not obvious.
Still, depending on the specific task at hand, the ideal architecture doesn’t always need to be the deepest or most evolved option. In some cases, these traditional methods continue to be valuable candidates for model design as they don’t require nearly as much data as deep learning models.
Another benefit of these earlier models is that they are “explainable” — the user knows exactly which features of the image participated in the prediction decision. For example, in one of our experiments, we are training a model that is using specific features describing the morphology of the oocyte, such as the relationship between the zona pellucida and the ooplasm.
Fast forward to the birth of Convolutional Neural Networks (CNNs)…
CNNs — which form a key component of our model — have truly revolutionized image classification and are considered state-of-the-art architecture for analyzing vast amounts of visual data.
Some key tasks that CNNs excel at include:
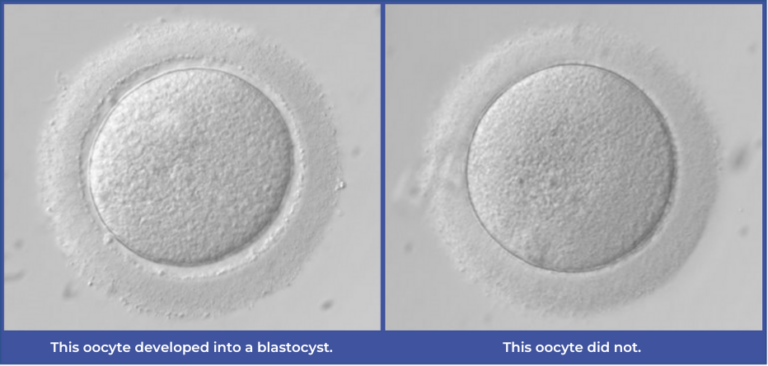
- • Image classification: The ability to tell the difference between two categories of images and assign a predicted output. For example, in our AI tool, this allows us to categorize an image of an oocyte that formed a blastocyst vs. an image that didn’t.
- As our model’s primary outcome is blastocyst formation, this capability is critical.
- • Object detection: The ability to identify which objects are present in an image and where they are located. Think about “object detection” as what the model sees in the image at a macro level. Each “object” is assigned a class label and the coordinates of that object are noted within the image. For us, this is useful as it can detect placement of the oocyte in a dish, ensuring we are capturing and analyzing the right data in the image.
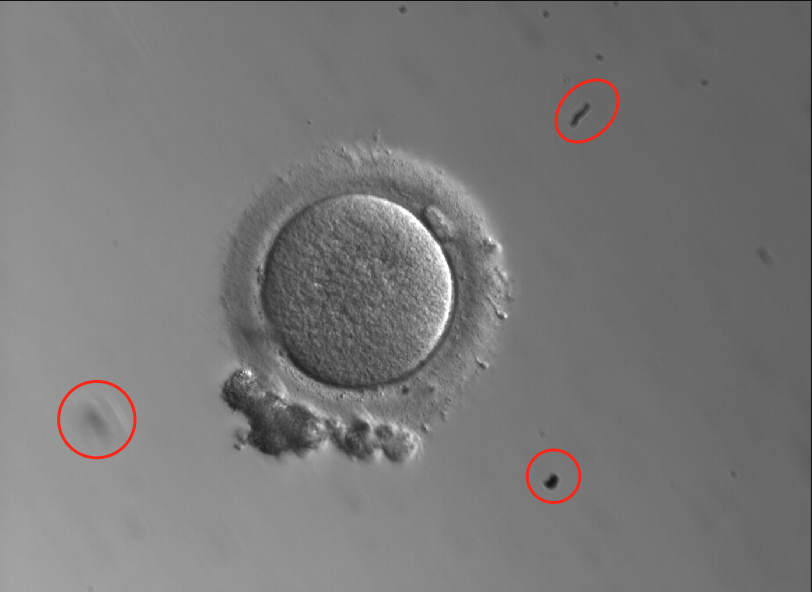
- Further to this, images may contain dirt from the microscope or other artifacts unique to a certain lab’s set-up. Our model needs to understand which part of the image is the oocyte versus other parts of the dish or artifacts, regardless of where in the image the oocyte happens to be sitting, to ensure that it focuses its analysis on the pixels that are relevant to the outcome predictions. Lastly, this also allows us the ability to get more granular to understand where specific morphological features are within the oocyte (polar body, zona pellucida, etc.).
- Here’s an example of an oocyte image containing debris within the dish:
- • Semantic segregation: While object detection takes a macro approach to image analysis, semantic segregation takes a micro approach. This is the ability to label each individual pixel in an image as belonging to one of the classes identified during the object detection exercise (for example, separating ooplasm vs. perivitelline space vs. zona pellucida).
- • Handling large quantities of data: Each pixel in an image is a data point that a CNN can use. Some high-quality images in our dataset have over 19 million pixels in a single image!
- CNN models have a number of tricks that allow them to take advantage of the spatial structure of images to reduce the complexity of the model and to learn meaningful visual features from the data. One critical example of this is the use of Efficient Training Algorithms based on stochastic gradient descent — an algorithm that has proven to be highly effective at training large-scale models on large-scale data.
- Since CNN models perform automatic feature detection based on importance for accurate prediction, they can find new features that the user is not aware of to make better predictions.
CNN models often require a massive number of data points to achieve high accuracy and generalization compared to classical machine learning techniques. In cases where your specialized dataset doesn’t include millions of images, a CNN model can first be pre-trained on a large, more general dataset like ImageNET and then fine-tuned on a smaller image dataset that’s directly relevant to the model’s task (i.e., oocyte images). This is called transfer learning and it’s proven to be a very useful and popular technique. In the pre-training phase, the goal is to get the model used to classifying broad categories of images by recognizing different outlines, textures or other simple features. Later on, when the smaller, task-specific dataset is introduced, the model transfers that broader classification knowledge to help focus its more nuanced training.
It’s clear from this example that any image classification model may have been trained on millions of images if you include the pre-training dataset. However, when you’re evaluating different models for your clinic, it’s important to understand how large the task-specific dataset is.
For example, our model was trained and tested using over 120,000 oocyte images – the largest dataset of its kind – to focus our model on identifying features that contribute to blastocyst formation.
For years, CNNs have been the go-to architecture for image classification and object detection tasks, but other architectures are quickly gaining steam in this space – most notably, Transformers.
ChatGPT, Apple Siri, Google Translate, and Amazon Alexa are all powered by transformer-based models. Transformer architectures were originally developed for natural language processing (NLP) tasks like understanding and generating text. However, researchers have found creative ways to apply them image analysis tasks, with unique advantages over CNNs.
CNNs focus first on understanding the important micro details of an image and then pan out to scan the image for patterns between these details. Transformers take a seemingly reverse approach where they evaluate the whole image at once by dividing it into smaller patches to understand the relationships between, and objects within, those patches. This holistic approach enables transformers to handle varying numbers of objects in an image, capture a more global context and better understand long-range dependencies between objects, which helps them detect objects and classify images accurately.
Vision Transformers (ViTs) and Detection Transformers (DETRs) have been found to achieve comparable or even superior performance to CNNs on large-scale image classification and object detection tasks, respectively. Both architectures continue to have their strengths and are used depending on the specific requirements of the task at hand.
Our team is always experimenting with the latest evolutions in deep learning architectures to develop new versions of our AI model that continuously improve our performance. These different architectures can be combined into an ensemble model to improve the predictive power of our tools.
New approaches that demonstrate potential model enhancement are carefully tested and assessed before incorporating them into our live model. Any model version that is used in your clinic through our software has been validated as the best-performing model at that time!
Just like building out a great team at your fertility clinic where each member contributes special skillsets, ensemble models can bring the best of different image classification and detection-focused architectures together to provide stronger predictions of an oocyte’s reproductive potential.
You Might Also Like …
Join our mailing list for dispatches on the future of fertility

